Tag: RNA-Seq
-
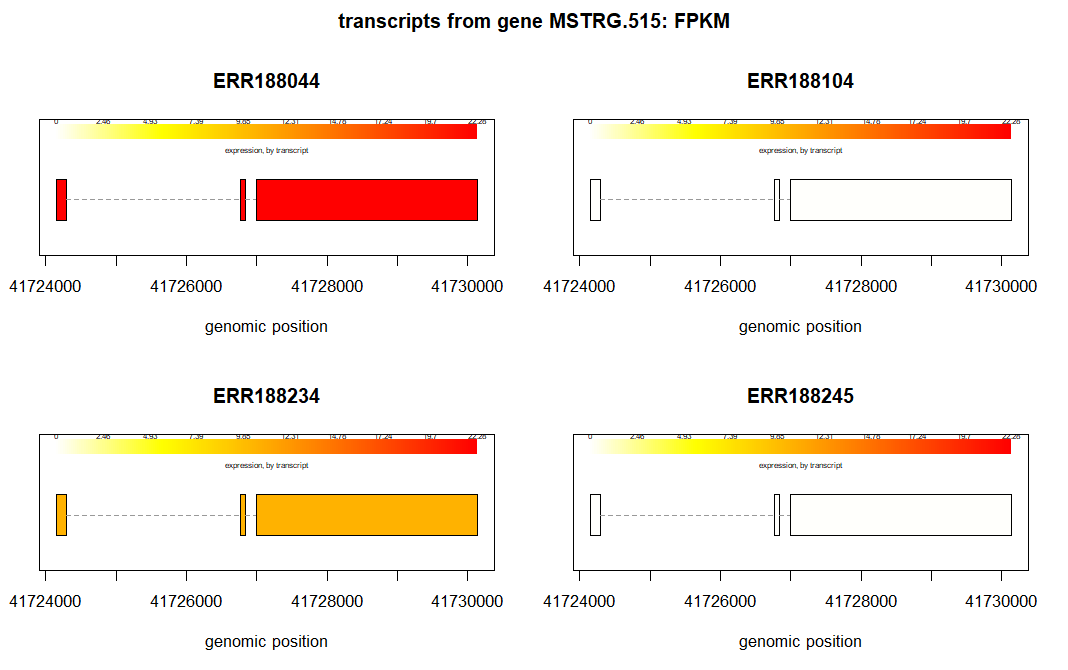
RNA-Seq Analysis in WSL – Part 3 : Differential expression analysis using Ballgown
Once we create the transcript abundance data using Stringtie, we can now perform the differential expression analysis. With Strigtie, we had created the estimated transcript abundance data which is compatible with Ballgown. The directory structure of the `for_ballgown` directory looks like this: To perform the differential gene expression analyis open Rstudio and change working directory…
-
RNA-Seq Analysis in WSL – Part 2 : Raw sequence reads to transcript abundance
In this post we will see how to use HISAT2 and Stringtie to align raw RNA-Seq reads to annotated genome and estimate transcript abundances.
-
RNA-Seq Analysis in WSL – Part 1 : Installation of tools
In this series of posts, we will see how we can perform RNA-Seq analysis in Windows operating system using linux tools in WSL. Also, the part of analysis which require “R”, will be done in the Windows system. We will be using following tools: Install samtools We need to install three things for this. Open…